
Xiaoyu Shi, Ph.D.
Assistant Professor, Department of Developmental and Cell Biology & Department of Chemistry
Ph.D., University of California, Davis
Postdoc, University of California, San Francisco
Research Areas: Super-Resolution Microscopy, Cell Biology, Spatial transcriptomics & proteomics, Cell Signaling, Single-Cell Analysis, Biophysics, Structural Biology, Systems Biology, Computational Microscopy.
Biosketch
Xiaoyu Shi is an Assistant Professor in the Department of Developmental and Cell Biology and the Department of Chemistry at the University of California, Irvine. Her research group uses optical and chemical approaches to develop super-resolution microscopy and spatial multiomics methods. She uses these cutting-edge technologies to study the molecular and cellular mechanisms of aging and cancers. Prior to UC Irvine, she received training in super-resolution microscopy and nonlinear optics during her postdoctoral research with Dr. Bo Huang at UCSF and her Ph.D. studies with Dr. Cheuk-Yiu Ng at UC Davis. Professor Shi is the recipient of the NIH Director’s New Innovator Award (DP2), the NIH Pathway to Independence Award (K99/R00), Chan Zuckerberg Initiative Frontiers of Imaging Award, and the Hellman Fellow.
Research Interests
Cells are the essential building blocks of life. They participate in the development and function of organs at all spatial scales – from molecular signaling to tissue patterning. The mission of Shi’s lab is to elucidate how cells drive developmental mechanisms across all scales. To answer these questions, we leverage super-resolution microscopy and chemical synthesis to develop novel cutting-edge imaging and multiomics methods and apply these methods to study the following specific topics:
I. How cellular machineries are organized at the molecular scale?
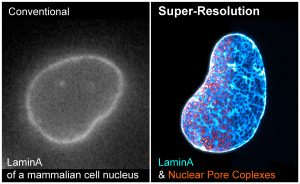 Seeing is believing. A major objective of our research program is to directly visualize how cells organize proteins, RNA, and DNA in space and time. We have developed a series of super-resolution microscopy methods to dissect the architecture of cellular structures at the molecular resolution, and with high labeling efficiency, brightness, and detection sensitivity. We are particularly interested in the structure-function relationships of the nuclear lamina, chromatin, cytoplasmic vesicles, and cellular protrusions called cilia.
Seeing is believing. A major objective of our research program is to directly visualize how cells organize proteins, RNA, and DNA in space and time. We have developed a series of super-resolution microscopy methods to dissect the architecture of cellular structures at the molecular resolution, and with high labeling efficiency, brightness, and detection sensitivity. We are particularly interested in the structure-function relationships of the nuclear lamina, chromatin, cytoplasmic vesicles, and cellular protrusions called cilia.
Methods: Expansion Microscopy (ExM), Stochastic Optical Reconstruction Microscopy (STORM), Structured Illumination Microscopy (SIM), Airyscan Microscopy
II. How do primary cilia mediate Hedgehog signaling?
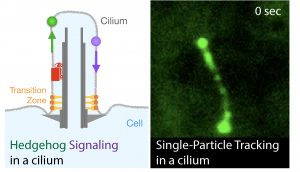 Hedgehog signaling pathway is the key signaling regulator of embryonic tissue patterning and adult tissue homeostasis. Defects in Hedgehog signaling can cause a multitude of developmental diseases, such as a congenital heart disease, and cancers, such as melanoma. Primary cilia, specialized protrusions on cell membrane, have an essential role in Hedgehog signaling in mammals. Our research objective is to determine the molecular mechanisms that underlie the regulation of Hedgehog signaling by primary cilia. Empowered by fast imaging and endogenous labeling technologies, we are able to directly watch and analyze the behavior of signaling and transporter molecules in action.
Hedgehog signaling pathway is the key signaling regulator of embryonic tissue patterning and adult tissue homeostasis. Defects in Hedgehog signaling can cause a multitude of developmental diseases, such as a congenital heart disease, and cancers, such as melanoma. Primary cilia, specialized protrusions on cell membrane, have an essential role in Hedgehog signaling in mammals. Our research objective is to determine the molecular mechanisms that underlie the regulation of Hedgehog signaling by primary cilia. Empowered by fast imaging and endogenous labeling technologies, we are able to directly watch and analyze the behavior of signaling and transporter molecules in action.
Methods: Live-cell single-particle tracking, Light-sheet microscopy, Total Internal Reflection Fluorescence Microscopy (TIRF), photoconversion
III. How single-cell multiomic data are organized at the spatial scale?
Over the last few years we are witnessing rapid advances in the fields of single-cell transcriptomics, genomics and proteomics. However, most tissues and organs are solid, with cells encased within a complex extracellular matrix network. Studying single cells from solid tissues usually requires their mechanical and enzymatic disaggregation. As a result, the positional information is lost, which is otherwise essential for proper understanding of how cells cooperate in tissues. We take optical and chemical approaches to develop novel, spatially-resolved GO3D multiomic technology, which isolates cells without homogenization, enables cell phenotyping with super resolution, and allows multiomics in the same single cells. The novel GO3D platform will significantly advance our knowledge on how cell phenotypes are maintained in native tissue and become perturbed in disease.
Awards and Honors
NIH Director’s New Innovator Award, NIH,2022-2027
Hellman Fellow, Hellman Foundation, 2022-2023
NIH Pathway to Independence Award, NIGMS,2018-2023
Frontiers of Imaging Award – Visual Proteomics Imaging , Chan Zuckerberg Initiative, 2021-2023
Selected Publications
- Label-retention expansion microscopy, X. Shi * #, Q. Li *, Z. Dai, A.A.Tran, S. Feng, A.D. Ramirez, Z. Lin, X. Wang, T.T. Chow, J. Chen D. Kumar, A. McColloch, J.F. Reiter, E.J. Huang, I.B. Seiple #, B. Huang #,
J. Cell Biol. 220 (9): e202105067 (2021). [link] **Image on Table of Content Page** - Polarized endosome dynamics engage cytoplasmic Par-3 that recruits dynein during asymmetric cell division. X. Zhao, J.Q. Garcia, K. Tong, X. Chen, B. Yang, Q. Li, Z. Dai, X. Shi, I.B. Seiple, B. Huang, S. Guo,
Science Advance, 7, eabg1244 (2021) [link] - Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules, A. Tulpule, J. Guan, D.S. Neel*, H.R. Allegakoen, Y.P. Lin, D. Brown, Y.-T. Chou, A. Heslin, N. Chatterjee, S. Perati, S. Menon, T.A. Nguyen, J. Debnath, A.D. Ramirez, X. Shi, B. Yang, S. Feng, S. Makhija, B. Huang#, T.G. Bivona#,
Cell, 184, 2649 (2021) [link] - A ciliopathy complex builds distal appendages to initiate ciliogenesis, D. Kumar, A. Rains, V. Herranz-Pérez, Q. Lu, X. Shi, D.L. Swaney, E. Stevenson, N.J. Krogan, B. Huang, C. Westlake, J.M. Garcia-Verdugo, B. Yoder, J.F. Reiter,
J. Cell Biol. 220 (9): e202011133 (2021). [link] - Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency, J. Zhang, D. Velmeshev, K. Hashimoto, Y.-H. Huang, J. W. Hofmann, X. Shi, J. Chen, A. M. Leidal, J. G. Dishart, M. K. Cahill, K. W. Kelley, S. A. Liddelow, W. W. Seeley, B. L. Miller, T. C. Walther, R. V. Farese Jr., J. P. Taylor, E. M. Ullian, B. Huang, J. Debnath, T. Wittmann, A. R. Kriegstein & E. J. Huang,
Nature, 588, 459 (2020) [link] - Nanotopography enhances dynamic remodeling of tight junction proteins through cytosolic complexes, X. Huang, X. Shi, M.E. Hansen, C.L. Nemeth, A. Ceili, B. Huang, T. Mauro, M. Koval, T.A. Desai,
ACS Nano, 14, 13192 (2020). [link] - X. Shi, G. Garcia, Y. Wang, J. Reiter, B. Huang, Alignment of super-resolution images for semi-flexible structures in 3D, PLoS One 0212735 (2019). [link]
- T.T. Chow, X. Shi, J.H. Wei, J. Guan, G. Stadler, B. Huang, E. Blackburn, Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection,
Nature Communications 9, 3583 (2018). [link] - X. Shi*, G. Garcia*, J.C. Van De Weghe, R. McGorty, G.J. Pazour, D. Doherty, B. Huang#, J.F. Reiter#, Super-resolution microscopy reveals that disruption of ciliary transition zone architecture is a cause of Joubert syndrome,
Nature Cell Biology 19, 1178 (2017). [link] **cover article** - J. Guan, H. Liu, X. Shi, S. Feng, B. Huang, Tracking multiple genomic elements using correlative CRISPR imaging and sequential DNA FISH,
Biophysical Journal 112,1077 (2017). [link] **New and Notable**



