
Research Interests
MAPK cascades in yeast and mammalian cells
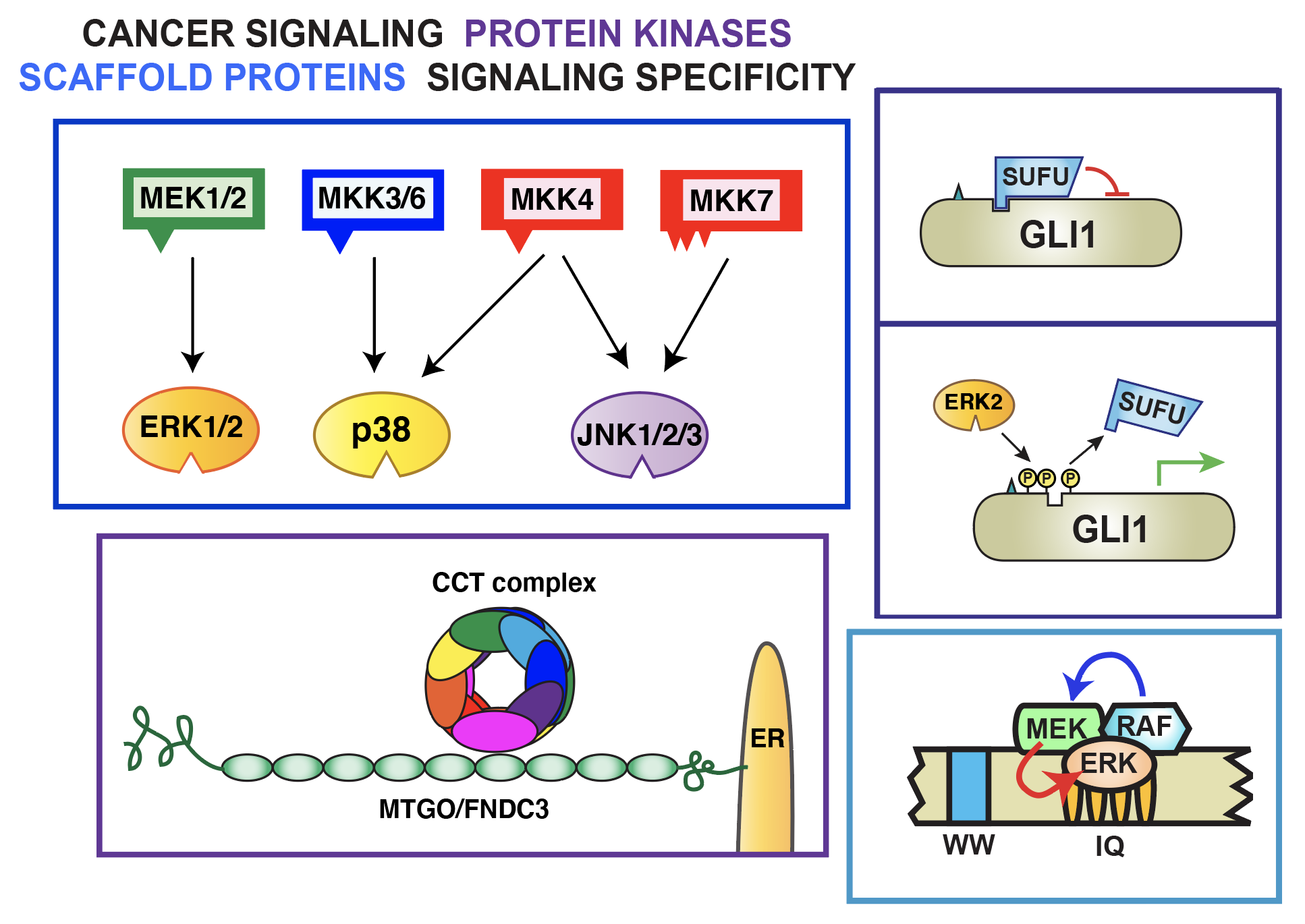
Cell Signaling – We study evolutionarily-conserved and disease-relevant cell signaling pathways, focusing on the following questions:
- How do protein kinases find their substrates? Can we use this knowledge to identify new substrates and regulatory strategies, and to find new ways to treat cancer and other diseases?
- How is specificity from signal to cellular response maintained when networks are highly interconnected, and different pathways use similar or overlapping components?
- What evolutionary logic underlies the structure of signaling and gene regulatory networks? Why are they so complicated and interconnected? What performance objectives might these designs achieve?
- How can we translate our increasingly sophisticated systems-level understanding of regulatory processes into new ideas for treating human disease?
We use an interdisciplinary approach, combining techniques from molecular cell biology, biochemistry and biophysics, genetics and genomics, and mathematical & computational biology and engineering to address these questions.
Our present emphasis is on mitogen-activated protein kinase (MAPK) signaling pathways. MAPK cascades participate in the regulation many biologically (and medically) important processes, including normal and pathological aspects of cell growth, division, differentiation, and death. The ubiquity and versatility of MAPK cascades make them ideal for addressing the questions we are interested in.
Recent Publications
- Bardwell AJ, Arif, U, andBardwell L (2026) AlphaFold3 predictions of novel GLI-SUFU interfaces identify binding-defective SUFU missense variants from medulloblastoma and Gorlin Syndrome patients. BioRxiv https://doi.org/10.64898/2026.01.06.698042
- Bheda P, Hou J, Aebersold R, Alon U, Bader JS, Bardwell L, Liu ET, Locke JCW, Mann M, Millar AJ, Naef F, Pilpel Y, Shamir R, Vitkup D (2025) Molecular Systems Biology at 20: reflecting on the past, envisioning the future.Mol Syst Biol. 2025 Dec;21(12):1667-1673. doi: 10.1038/s44320-025-00170-w.
- Lara-Gonzalez, P, Variyar S*, Moghareh, S*, Nguyen, ACN*, Kizhedathu, A, Budrewicz, J, Schlientz, A, Varshney, N, Bellaart, A, Oegema, K, Bardwell, L and Desai, A (2024) Cyclin B3 is a dominant fast-acting cyclin that drives rapid early embryonic mitoses, Journal of Cell Biology 223 (11): e202308034.
- Press Release: Unlocking the secrets of rapid cell division
- Bardwell, L, and Thorner, J (2023) Mitogen-activated protein kinase (MAPK) cascades–A yeast perspective. Enzymes. 54:137-170.
- Bardwell AJ, Paul, M, Yoneda, KC, Andrade-Ludena, MD, Nguyen, OT, Fruman, DA, and Bardwell L (2023) The WW domain of IQGAP1 binds directly to the p110α catalytic subunit of PI 3-kinase, Biochemical Journal, 480 (10): 729–750. (Cover Article)
- Bardwell AJ, Wu, B, Sarin KY, Waterman ML, Atwood SX, and Bardwell L (2022) ERK2 MAP kinase regulates SUFU binding by multisite phosphoryaltion of GLI1. Life Science Alliance 5(11):e202101353 DOI: 10.26508/lsa.202101353.
- Press Release: How a protein breaks free to cause deadly cancers
- Han H, Nakaoka HJ, Hofmann L, Zhou JJ, Yu C, Zeng L, Nan J, Seo G, Vargas RE, Yang B, Qi R, Bardwell L, Fishman DA, Cho KWY, Huang L, Luo R, Warrior R, Wang W. (2022) The Hippo pathway kinases LATS1 and LATS2 attenuate cellular responses to heavy metals through phosphorylating MTF1. Nature Cell Biology 24:74-87. doi: 10.1038/s41556-021-00813-8.
- Bardwell L (2020) Cancer Mutations: Molecular MEKansims. Current Biology 30:R222–R224
- Lagunes, L, Bardwell L and Enciso, GA. (2020) Effect of magnitude and variability of energy of activation in multisite ultrasensitive biochemical processes. PLoS Computational Biology, 16:e1007966.
- Bardwell L (2019) Pseudokinases: Flippping the ATP for AMPylation. Current Biology 29:R16–R37
- Syed, A, Lukacsovich, T, Pomeroy, M, Bardwell AJ, … Bardwell L, Marsh, JL, MacGregor, GR (2019) Miles to go (mtgo) encodes FNDC3 proteins that interact with the chaperonin subunit CCT3 and are required for NMJ branching and growth in Drosophila. Developmental Biology, 445: 37-53
- Bardwell, AJ, Lagunes, L, Zebarjedi, R, and Bardwell, L (2017) The WW domain of the scaffolding protein IQGAP1 is neither necessary nor sufficient for binding to the MAPKs ERK1 and ERK2. J. Biol. Chem., 292: 8750-8761
- Bardwell AJ and Bardwell, L (2015) Two hydrophobic residues can determine the specificity of MAP kinase docking interactions. J. Biol. Chem., 290:26661-266674
- Mattson-Hoss MK, Niitani Y, Gordon EA, Jun Y, Bardwell, L, Tomishige M, and Gross SP (2014) CK2 activates kinesin via induction of a conformational change. Proc Natl Acad Sci USA 2014 111:7000-7005.
- Gordon EA, Whisenant, TC, Zeller, M, Kaake, RM, Gordon, WM, Krotee, P, Patel, V., Huang, L, Baldi, P and Bardwell, L (2013), Combining docking site and phosphosite predictions to find new substrates: Identification of Smoothelin-like-2 (SMTNL2) as a c-Jun N-terminal kinase (JNK) substrate. Cellular Signalling, 25:2518-2529.
- Chan, C, Liu, X, Wang, L, Bardwell, L, Nie, Q, Enciso, G (2012) Protein Scaffolds Can Enhance the Bistability of Multisite Phosphorylation Systems. PLoS Computational Biology 8(6): e1002551.
- Xu, J, Reddy, Anand, P, Shu, Z, Cermelli, S, Mattson, MK, Tripathy, SK, Hoss, MT, James, NS, King, SJ, Huang, L, Bardwell, L and Gross, SP (2012) Casein Kinase 2 Reverses Tail-Independent Inactivation of Kinesin-1. Nature Communications, 3:754.
- Chou, C-S, Bardwell, L, Nie, Q and Yi, TM (2011) Noise filtering tradeoffs in spatial gradient sensing and cell polarization response. BMC Systems Biology, 5:196.
- Bernardo, AS, Faial, T, Gardner, L, Niakan, KK, Ortmann, D, Senner, CE, Callery, EM, Trotter, MW, Hemberger, M, Smith, JC, Bardwell, L, Moffett, A, Pedersen, RA (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell, 9:144-55.
- Bardwell, L (2011) Synthetic Biology: Modulating the MAP kinase module. Current Biology, 21:R249-51.
- Whisenant, TM, Benz, R, Ho, DM, Rogers, JM, Kaake, RM, Gordon, EA, Huang, L, Baldi, P, and Bardwell, L (2010) Computational prediction and experimental verification of novel MAP kinase substrates including Gli transcription factors. PLoS Computational Biology, 6:e10009084.
- Haney, S., Bardwell, L+, and Nie, Q+ (2010) Ultrasensitive responses and specificity of cell signaling. BMC Systems Biology, 4:119.
- Liu, X, Bardwell, L+ and Nie, Q+ (2010) A combination of multisite phosphorylation and substrate sequestration produces switch-like responses. Biophysical Journal, 98:1396-407.
- Bardwell, AJ, Frankson, E and Bardwell, L (2009) Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 284:13165-73.
* = equal contribution; + = co-corresponding authors.
