Website updates: 09/2020

Research Topics: Spatial and Temporal Regulation of Gene Expression and Chromatin Landscape During Embryogenesis
Total Peer Reviewed Publication: 115 papers
Citations: 9521
h-index: 49
i10 index 77
http://www.kencho-lab.org
kwcho@uci.edu

Overall Goal:
A central question in biology is to understand how the naive genome in the early embryo progressively undergoes a series of modifications to control gene expression in “time and space” such that proper cellular differentiation programs are correctly implemented as cells differentiate from pluripotent to specific cell types. Our goal is to uncover the integrative roles of maternal TFs in regulating the onset of zygotic genome activation (the first phase of gene expression in embryos), coordinating nucleosome phasing and histone modifications on target genes, and shaping the 3D architecture of chromatin. We combine both genomic and imaging approaches to provide important insights into the unifying principles that drive genome activation.
Transcriptomics and Epigenomics
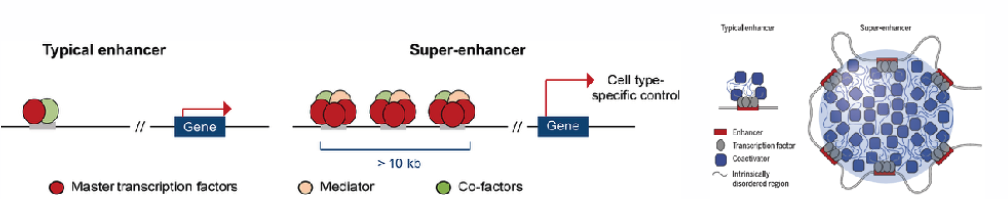
- Zygotic genome activation is a critical period in metazoan development whereby the control of cell fates is transferred from maternal genes to zygotic genes. We examine how the interplay between maternal transcription factor and epigenetics play during zygotic genome activation, which later results in the establishment of novel specific subsets of enhancers called super enhancer which concentrate the transcription apparatus and form phase-separated multimolecular assemblies in the nucleus.
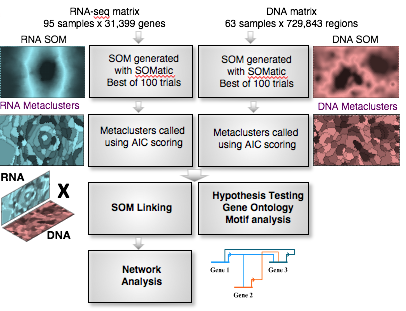
- A major unanswered question in biology is how differentiation of the myriad cell types of the adult body is hardwired in the genome. Using the frog and mouse as model systems, our goal is to elucidate the gene regulatory mechanisms controlling endoderm formation by combining experimental and computational approaches. We analyze high-dimension genomic data sets originating from dozens of RNA-seq, ChIP-seq and ATAC-seq datasets, and perform computational network modeling to infer the endoderm gene regulatory network.
Live Imaging During Early Embryogenesis
- Recent work revealed that super enhancers (SE) concentrate the transcription apparatus, and form phase-separated multimolecular assemblies, that can be visualized as discrete punctae in the nuclei of cells. Our hypothesis is that SEs associated with maternal TFs form dynamic, long-range chromatin looping complexes and are components of nuclear condensates. Our interest is to examine how maternal factors regulate 3D genome conformation and what components drive formation and dissolution of transcriptional condensates.
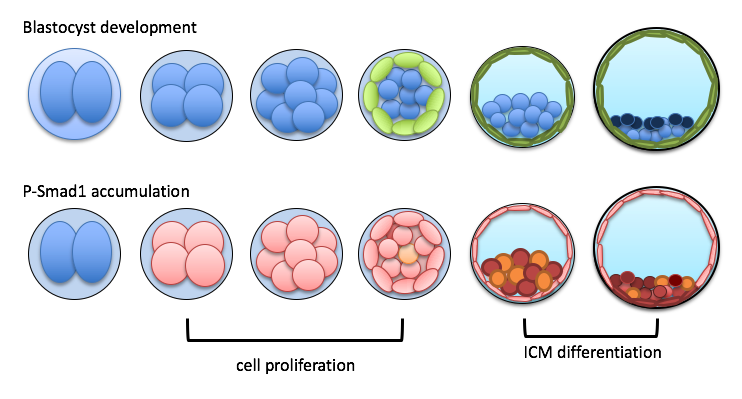
- Implantation occurs at the very early stage of pregnancy, at which the dividing mammalian embryo adheres to the wall of the uterus. We have previously shown that Bone Morphogenetic Proteins (BMPs) regulates the cell division rate and are key growth factors during this process. We apply high level confocal imaging and genomic tools at the single cell level to understand how BMP signaling regulate the process. In addition, we also developed a method to assess the quality of preimplantation embryos using the phasor-FLIM(Fluorescence Lifetime Imaging Microscopy) method, which is a non-invasive live imaging approach to capture endogenous autofluorescent metabolic markers to distinguish the quality of pre-implantation embryos. Our work will shed light on an issue of early pregnancy loss: failure of the embryos to undergo normal development.
Genome engineering
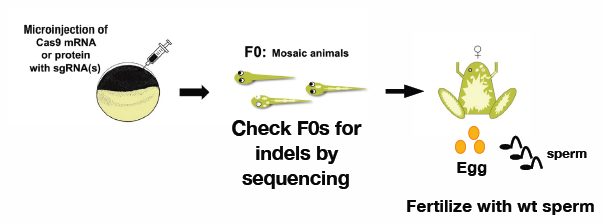
- The CRISPR/Cas9 system offers a significant advantage over other systems for examining mutations in the F0 generation in non-mammalian systems. We use Xenopus tropicalis to develop methods to eliminate a large genomic region and induce a homologous recombination at a desired locus within the genome.
Recent Publication:
2020
Mukherjee S, Chaturvedi P, Rankin SA, Fish MB, Wizla M, Paraiso KD, MacDonald M, Chen X, Weirauch MT, Blitz IL, Cho KWY, Zorn AM. Sox17 and b-catenin co-occupy Wnt-responsive enhancers to govern the endodermal gene regulatory network. 2020. eLife, in press. (BioRxiv doi: https://doi.org/10.1101/2020.02.19.956565)
Afouda BA, Nakamura Y, Shaw S, Charney RM, Paraiso KD, Blitz IL, Cho KWY, Hoppler S. Foxh1/Nodal Defines Context-Specific Direct Maternal Wnt/β-Catenin Target Gene Regulation in Early Development iScience 2020 Jul 24;23(7):101314.
Gilchrist MJ, Veenstra GJC, Cho KWY. Transcriptomics and Proteomics Methods for XenopusEmbryos and Tissues. Cold Spring Harb Protoc. 2020 Feb 3;2020(2):pdb.top098350.
Gilchrist MJ, Cho KWY, Veenstra GJC. Genomics Methods for Xenopus Embryos and Tissues. Cold Spring Harb Protoc. 2020 (5):pdb.top097915. doi: 10.1101/pdb.top097915.
2019
Paraiso KD, Blitz IL, Zhou JJ, Cho KWY. Morpholinos Do Not Elicit an Innate Immune Response during Early Xenopus Embryogenesis. Dev Cell. 2019 49:643-650.e3.
Paraiso KD, Blitz IL, Coley M, Cheung J, Sudou N, Taira M, Cho KWY. Endodermal Maternal Transcription Factors Establish Super-Enhancers during Zygotic Genome Activation. Cell Rep. 2019 27:2962-2977.e5.
Cho JS, Blitz IL, Cho KWY. DNase-seq to Study Chromatin Accessibility in Early Xenopus tropicalis Embryos. Cold Spring Harb Protoc. 2019 (4).
Ma N, Mochel NR, Pham PD, Yoo TY, Cho KWY*, Digman MA*. Label-free assessment of pre-implantation embryo quality by the Fluorescence Lifetime Imaging Microscopy (FLIM)-phasor approach. Sci Rep. 2019, 9:13206.
2017:
Charney, R.M., Forouzmand, E., Cho, J.S., Cheung, J., Paraiso, K.D., Yasuoka, Y., Takahashi, S., Taira, M., Blitz, I.L., Xie, X. and. Cho K.W.Y. (2017). Foxh1 occupies cis-regulatory modules prior to dynamic transcription factor interactions controlling the mesendoderm gene program. Dev Cell. 40:1-13.
Charney RM, Paraiso KD, Blitz IL, Cho KWY. (2017). A gene regulatory program controlling early Xenopus mesendoderm formation: Network conservation and motifs. Semin Cell Dev Biol. 66:12-24.
Holmes, W.R., Mochel, S., Wang, Q., Du, H., Cinquin, O., Cho, K.W*., Nie, Q*. *contributed equally, co-senior authors. (2017). Intracellular noise aids construction of early embryonic structures. PLoS Comput Biol. 13(1):e1005320.
2016:
Owens, NL, Blitz, IL, Lane, MA, Overton, JD, Gilchrist, MJ#, Cho, KWY#., Khokha, MK # (2016). Embryogenesis kinetics measured by high-resolution absolute quantitation of transcripts. 1: co-first authros, contributed equally. # co-senior authors. Cell Reports. 14, 632-647.
Blitz, IL, Fish, MB, and Cho, KWY. (2016). Leapfrogging: Primordial Germ Cell Transplantation Permits Recovery of CRISPR/Cas9-Induced Mutations in Essential Genes. Development, 143:2868-75. PMID: 27385011
Blitz, IL, Paraiso, KD, Patrushev, I, Chiu, WTY, Cho, KWY*, Gilchrist,MJ.* (2016). . *contributed equally, co-senior authors. A catalog of Xenopus tropicalis transcription factors and their regional expression in the early gastrula stage embryo. Dev Biol, 2016 S0012-1606(16) 30118.


